If the search is finished, an output report is shown. The report for MS/MS data is shown below. For other data, see Dereplication in MS Data, Dereplication in LC-MS Data, and Dereplication in MSI Data.
Peptide sequence candidates in yellow rows contain a regular expression used in Searched Sequence. Note that the regular expression may be matched in any linearized sequence of a cyclic, branched, branch-cyclic peptide sequence or cyclic polyketide.
The table can be exported into a csv file using the command 'File -> Export to CSV' and then imported, e.g., to MS Excel. It can be also exported into a HTML page using the command 'File -> Export to HTML' including details about peptide sequence candidates. The report can be saved using the command 'File -> Save Results...' and re-opened using the command 'File -> Open Results...' (the files created using an older version of the software cannot be opened in a newer version).
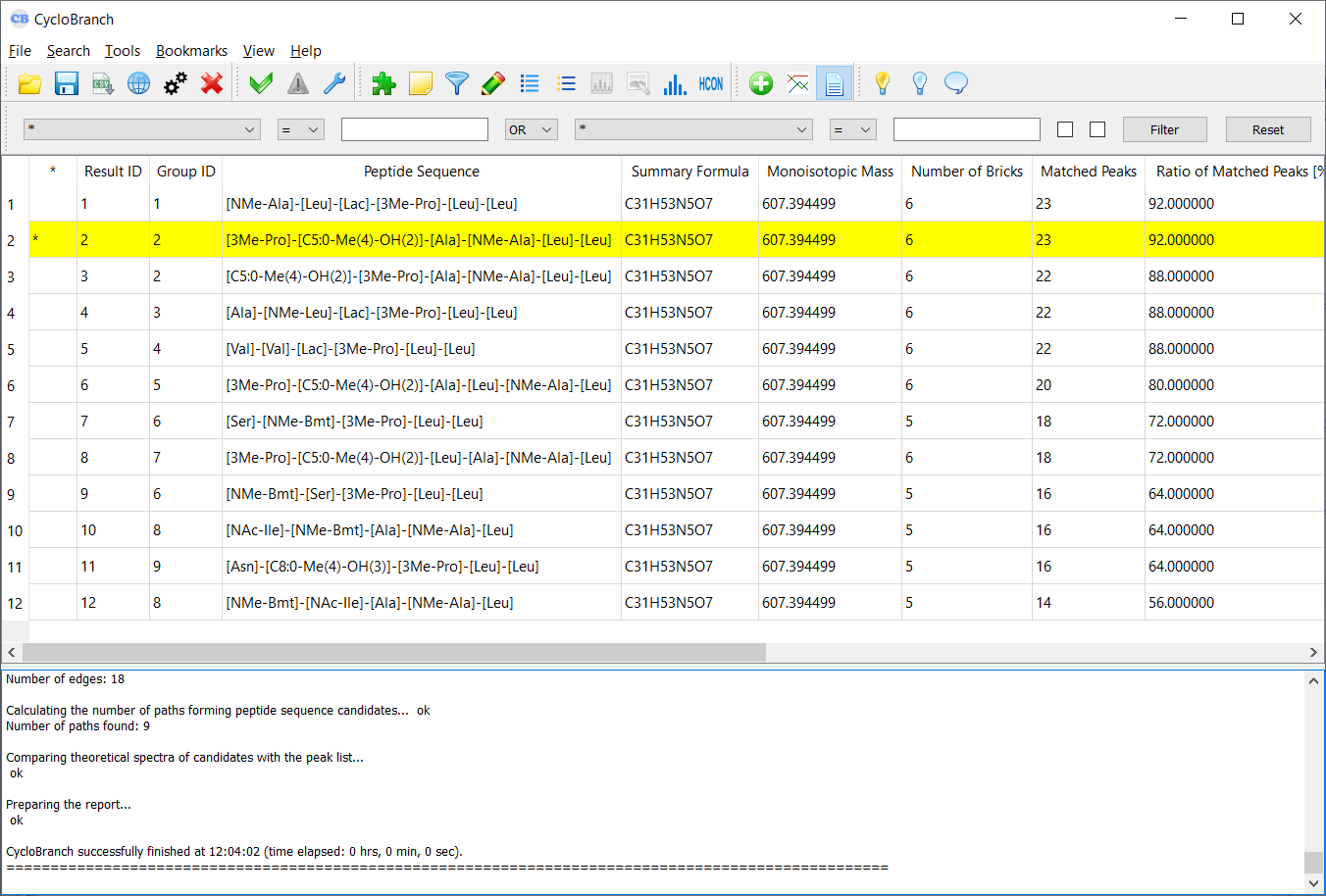
De novo sequencing - example of output report.
The report contains the following columns when a peptide is being searched:
-
* - it indicates that a row is highlighted; you can sort the list by this column to see all highlighted rows at the top
-
Result ID - a rank of a peptide sequence candidate according to a score defined in Score Type (if the mode "De Novo Search Engine" or "Compare Peaklist with Database - MS/MS data" is used; see Mode)
-
Group ID - an identifier of a group of peptide sequence candidates generated from the same path in a de novo graph (if the mode "De Novo Search Engine" is used; see Mode)
-
Name - a name of a peptide (if the mode "Compare Peaklist with Database - MS/MS data" is used; see Mode)
-
Scan ID - an id of a scan which is used as Scan no. (if the mode "Compare Peaklist(s) with Spectrum of Searched Sequence" is used; see Mode)
-
Title - a title of an experimental spectrum (if the mode "Compare Peaklist(s) with Spectrum of Searched Sequence" is used; see Mode)
-
Peptide Sequence - a peptide sequence candidate
-
Summary Formula - a summary formula of a neutral molecule. When a sequence tag is reported, the formula is incomplete. For example, a formula "C38H69N7O7 (partial)" of a sequence tag "[198.136914]-[Val/Ival]-[Me-Leu]-[Ala/bAla/Sar]-[Ala/bAla/Sar]-[Me-Leu]-[Me-Leu]-[Leu/Ile/NMe-Val]-[268.179095]" does not include the unknown blocks [198.136914] and [268.179095].
-
Monoisotopic mass - a monoisotopic mass of a neutral modlecule
-
Number of Bricks - a number of building blocks in a peptide sequence candidate
-
Matched Peaks - a number of matched peaks between an experimental spectrum and a theoretical spectrum of a peptide sequence candidate
-
Ratio of Matched Peaks [%] - a ratio of matched peaks to all peaks in an experimental spectrum
-
Sum of Relative Intensities - a sum of relative intensities of matched peaks
-
Weighted Ratio of Matched Peaks - a sum of relative intensities of matched peaks / a sum of relative intensities of matched and unmatched peaks
-
Cosine Similarity
-
A - the number of a-ions
-
B - the number of b-ions
-
C - the number of c-ions
-
X - the number of x-ions
-
Y - the number of y-ions
-
Z - the number of z-ions
Note that the numbers of fragment ions are reported if a desired type of a fragment ion has been selected in Ion Types in Theoretical Spectra. The different types of fragment ions are reported for linear and cyclic polyketides, see Nomenclature of Linear Polyketide Series for details.
The following additional columns are shown if a linear or branched peptide was searched:
-
N-terminal Modification - N-terminal modification of the peptide
-
C-terminal Modification - C-terminal modification of the peptide
The following additional column is shown if a branched or branch-cyclic peptide was searched:
-
Branch Modification - a terminal modification of a branch
The following additional column is shown if a cyclic peptide was searched:
-
Scrambled Peaks - a number of matched scrambled peaks between an experimental spectrum and a theoretical spectrum of a peptide sequence candidate
The following additional columns are shown if a linear polyketide was searched:
-
Left Terminal Modification - a terminal modification (N- or C-terminal)
-
Right Terminal Modification - a terminal modification (N- or C-terminal)
The following columns are shown if MS, LC-MS, and MSI data is processed (see Mode):
-
*
-
Spectrum ID - a number an experimental spectrum when a series of peaklists was processed (e.g., LC-MS or MSI data)
-
Title - a title of a spectrum
-
Matched Peaks
-
Ratio of Matched Peaks [%]
-
Sum of Relative Intensities
-
Weighted Ratio of Matched Peaks
The following column is shown if the LC-MS data is processed:
-
Time - retention time (time units are dataset dependent)
The following columns are shown if the imaging mass spectra are processed:
-
Coordinate X - x coordinate of a single mass spectrum
-
Coordinate Y - y coordinate of a single mass spectrum
Details about a peptide sequence candidate are shown after a double click (or Enter key press) on a row in the output report. See the details on extra pages.
Linear Peptide Detail Window
Cyclic Peptide Detail Window
Branched Peptide Detail Window
Branch-cyclic Peptide Detail Window
Linear Polyketide Detail Window
Cyclic Polyketide Detail Window
Dereplication in MS Data
Dereplication in LC-MS Data
Dereplication in MSI Data
